Abstract
Introduction:
It is well established that genetic prognostication using fluorescence in situ hybridization (FISH) for the common recurrent abnormalities deletion 13q, trisomy 12, deletion 11q, and deletion 17p hierarchically stratifies time to first treatment (TTFT) and overall survival (OS) for chronic lymphocytic leukemia (CLL) patients (pts). Approximately 20% of CLL pts are negative for each of these abnormalities (normal 12/13/11/17), which is considered a favorable prognostic finding. Although favorable, outcomes within this group are heterogeneous, with limited information as to the importance of other simultaneous factors. Limited studies have shown a subset of these pts have complex karyotypes, suggesting standard CLL FISH may be insufficient for prognostication of this group, prompting a need for studies focusing specifically on this subset of pts (Haferlach et. al. Leukemia 2007; Baliakas et. al. Blood 2019). Here, we investigated the normal 12/13/11/17 pts to interrogate the utility of additional genetic testing and identify prognostically important variables.
Methods:
We conducted a retrospective analysis of all treatment-naïve CLL pts with cytogenetics performed at The Ohio State University Cytogenetics Laboratory from 2008 to 2018 for whom standard 12/13/11/17 CLL FISH were normal on their first cytogenetic analysis at our institution. Chromosome banding analysis was performed on cells stimulated with CpG oligonucleotides, phorbol-12-myristate-13-acetate, and pokeweed mitogen and analyzed according to standard laboratory procedures. FISH using probes for the standard CLL panel, D13S319, D12Z3 , ATM, TP53, and an expanded panel including BCL6, MYC, REL, SEC63/MYB, CDKN2A, and IGH were done according to manufacturer's recommendations. PCR was used to determine IGHV mutational status. TTFT and OS were both measured from diagnosis; pts not starting treatment were censored at death or last follow-up for TTFT, and pts who were alive were censored at last follow-up for OS. Cox regression models were used to evaluate TTFT and OS. A multiple imputation procedure was used to impute missing data and obtain regression estimates from combining results across 30 imputed datasets.
Results:
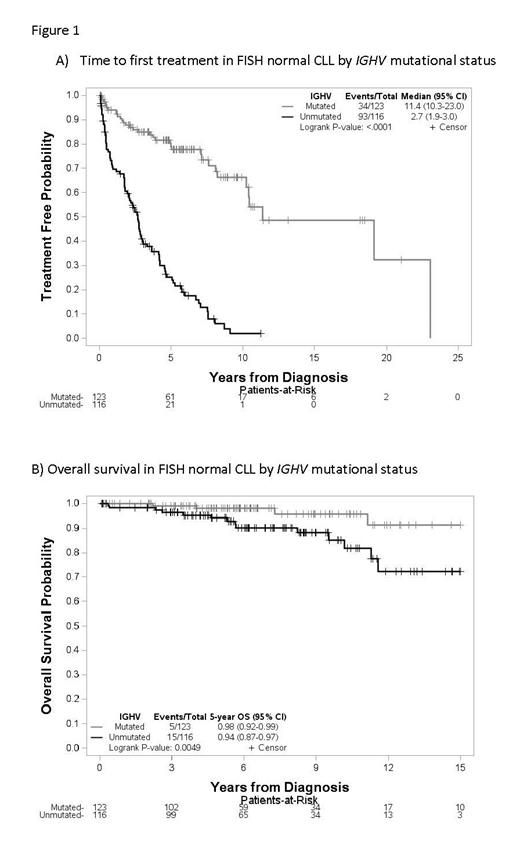
We analyzed 280 pts with a median follow up of 6.1 years among survivors since diagnosis. Median age at diagnosis was 58, and 63% were men. 95% of pts had low to intermediate Rai stage at diagnosis, 49% were IGHV unmutated, and 6% expressed IGHV3-21. 35% of pts showed an abnormal karyotype, with 8% being complex (3 or more abnormalities). Additional FISH testing showed 7% had deletion of IGH, 5% had BCL6 gain/rearrangement, 5% had deletion 6q, 4% had gain/rearrangement of MYC, 4% had gain of REL (limited subset tested, n=180), and 3% had IGH rearrangement. None of the tested pts (limited subset tested, n=115) had CDKN2A deletion. 144 pts (51%) progressed to requiring treatment, with a median TTFT of 6.5 years (95% CI 5-7.6). On univariable analysis, male sex, advanced Rai stage, increasing Beta-2-microglobulin (B2M), unmutated IGHV, increasing karyotypic complexity, gain of REL, and deletion 6q by FISH were significant predictors of shorter TTFT. However, only increasing B2M (p=0.03, HR 1.15 (95% CI 1.01-1.32)) and unmutated IGHV (p<0.0001, HR 5.84 (95% CI 3.83-8.89)) retained significance in a multivariable model. In terms of OS, there were only 24 deaths in the study cohort. Univariable analysis found age at diagnosis, increasing B2M, unmutated IGHV, and gain of REL statistically significant predictors of worse OS. Older age (p<0.0001, HR 1.56 (95% CI 1.26-1.94)) and unmutated IGHV (p=0.0037, HR 5.45 (95% CI 1.75-17.00)) retained significance in the multivariable model. Richter transformation occurred in 7 pts (2.5%), 4 with unmutated IGHV, 1 with mutated IGHV, and two with unknown IGHV status, with a median time from diagnosis to transformation of 9.5 years. Notably, for those pts with normal 12/13/11/17 FISH and mutated IGHV, median TTFT was 11.4 years (95% CI 10.3-23.0, Figure 1A) and OS at 5 years was 98% (95% CI 92-99%, Figure 1B).
Conclusions:
Additional cytogenetic testing demonstrates alternative genetic alterations are common in normal 12/13/11/17 CLL pts, however, our data do not show that these alterations are significant independent predictors of TTFT or OS. While normal FISH CLL is a relatively heterogeneous group, pts with mutated IGHV represent a subset of pts with very low risk disease.
Ruppert: Telios Pharma: Consultancy. Byrd: Novartis, Trillium, Astellas, AstraZeneca, Pharmacyclics, Syndax: Consultancy, Honoraria; Vincerx Pharmaceuticals: Current equity holder in publicly-traded company, Membership on an entity's Board of Directors or advisory committees; Newave: Membership on an entity's Board of Directors or advisory committees. Bhat: Beigene: Consultancy; Aptitude Health: Honoraria; Onclive: Honoraria; AstraZeneca: Consultancy. Kittai: Janssen: Consultancy; Abbvie: Consultancy; Bristol-Meyers Squibb: Consultancy. Rogers: Acerta Pharma: Consultancy; Pharmacyclics LLC: Consultancy; Innate Pharma: Consultancy; Genentech: Consultancy, Research Funding; AstraZeneca: Consultancy; Janssen Pharmaceuticals, Inc: Research Funding; AbbVie Inc.: Consultancy, Research Funding; ovartis Pharmaceuticals Corporation: Research Funding. Woyach: AbbVie Inc, ArQule Inc, AstraZeneca Pharmaceuticals LP, Janssen Biotech Inc, Pharmacyclics LLC, an AbbVie Company,: Consultancy; AbbVie Inc, ArQule Inc, Janssen Biotech Inc, AstraZeneca, Beigene: Other: Advisory Committee; AbbVie Inc, Loxo Oncology Inc, a wholly owned subsidiary of Eli Lilly & Company: Research Funding; Gilead Sciences Inc: Other: Data & Safety.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal